Periodic boundary conditions#
boundary value problem on \(\textbf{x}\in\Omega\)
\[\nabla^2 u = f(\textbf{x})\]
Neumann boundary condition on \(\textbf{x}\in\partial\Omega_{\text{D}}\)
\[u = u_{\text{D}}\]
Neumann boundary condition on \(\textbf{x}\in\partial\Omega_{\text{N}}\)
\[\textbf{n}\cdot\nabla u = u_{\text{N}}\]
periodic boundary condition on \(\textbf{x}\in\partial\Omega_{\text{M}}\cup\in\partial\Omega_{\text{S}}\)
\[u\vert_{\partial\Omega_{\text{M}}} = u\vert_{\partial\Omega_{\text{S}}}\]
import numpy as np
from dolfinx.fem import FunctionSpace
from lucifex.mesh import rectangle_mesh, mesh_boundary
from lucifex.fem import Function
from lucifex.solver import bvp, BoundaryConditions, OptionsPETSc
from lucifex.viz import plot_colormap, plot_line
from lucifex.utils import cross_section
from lucifex.pde.poisson import poisson
Lx = 2.0
Ly = 1.0
mesh = rectangle_mesh(Lx, Ly, 64, 64)
boundary = mesh_boundary(
mesh,
{
"left": lambda x: x[0],
"right": lambda x: x[0] - Lx,
"lower": lambda x: x[1],
"upper": lambda x: x[1] - Ly,
},
)
def right_to_left(x: np.ndarray) -> np.ndarray:
return np.vstack((x[0] - Lx, x[1]))
def upper_to_lower(x: np.ndarray) -> np.ndarray:
return np.vstack((x[0], x[1] - Ly))
fs = FunctionSpace(mesh, ('P', 1))
Periodic in the \(x\)-direction (\(\partial\Omega_{\text{D}}\cup\partial\Omega_{\text{N}}\neq\varnothing\))#
\[c(x=0,y)=c(x=L_x,y)\]
\[c(x,y=0)=c(x,y=L_y)=0\]
bcs_pbc_x = BoundaryConditions(
('periodic', boundary['right'], right_to_left),
("dirichlet", boundary['lower', 'upper'], 0.0),
)
f = Function(fs, lambda x: np.cos(2 * np.pi * x[0] / Lx) * np.cos(2 * np.pi * x[1] / Ly), name='f')
u = Function(fs, name='u')
u_solver = bvp(poisson, bcs_pbc_x)(u, f)
u_solver.solve()
plot_colormap(u)
(<Figure size 640x480 with 2 Axes>, <Axes: xlabel='$x$', ylabel='$y$'>)
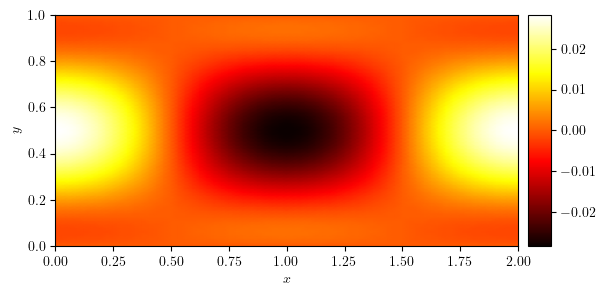
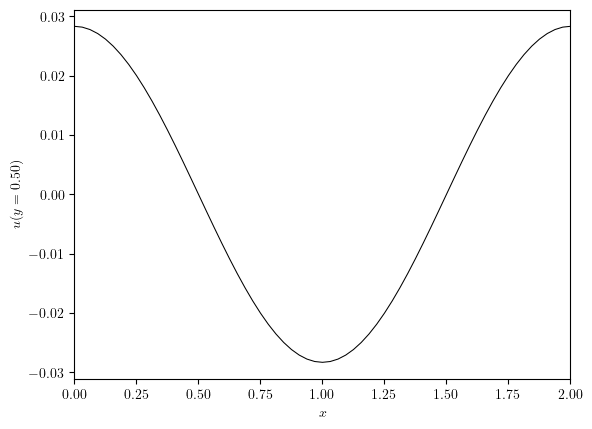
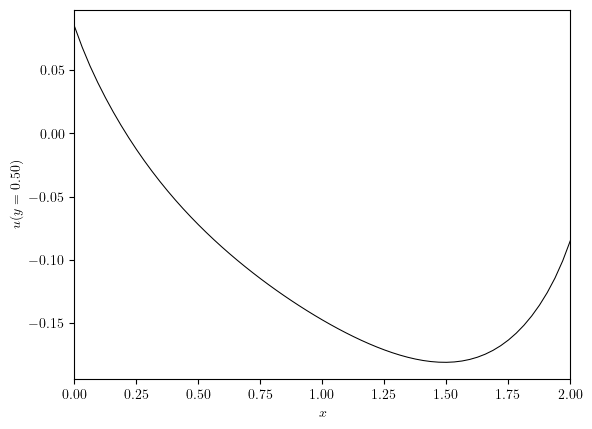
x_axis, ux, y_value = cross_section(u, 'y', 0.5)
plot_line((x_axis, ux), x_label='$x$', y_label=f'${u.name}(y={y_value:.2f})$')
(<Figure size 640x480 with 1 Axes>, <Axes: xlabel='$x$', ylabel='$u(y=0.50)$'>)
Periodic in the \(x\) and \(y\) directions (\(\partial\Omega_{\text{D}}\cup\partial\Omega_{\text{N}}=\varnothing\))#
\[c(x=0,y)=c(x=L_x,y)\]
\[c(x,y=0)=c(x,y=L_y)\]
bcs = BoundaryConditions(
('periodic', boundary['upper'], upper_to_lower),
('periodic', boundary['right'], right_to_left),
)
u = Function(fs, name='u')
f = Function(fs, lambda x: np.cos(2 * np.pi * x[0] / Lx) * np.cos(2 * np.pi * x[1] / Ly), name='f')
u_solver = bvp(poisson, bcs)(u, f)
u_solver.solve()
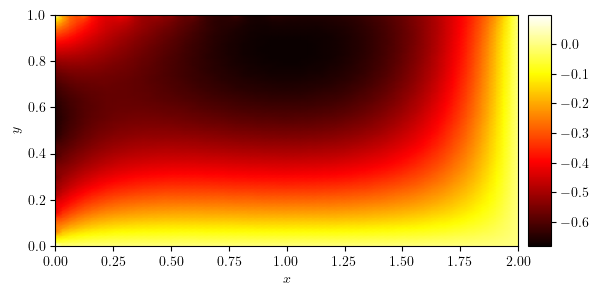
plot_colormap(u)
(<Figure size 640x480 with 2 Axes>, <Axes: xlabel='$x$', ylabel='$y$'>)
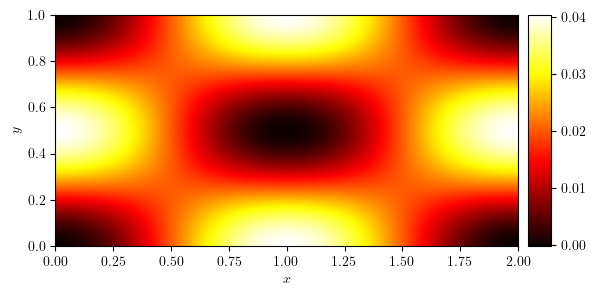
Antiperiodic in the \(x\)-direction#
\[c(x=0,y)=-c(x=L_x,y)\]
\[c(x,y=0)=c(x,y=L_y)=0\]
bcs = BoundaryConditions(
('antiperiodic', boundary['right'], right_to_left),
("dirichlet", boundary['lower', 'upper'], 0.0),
)
f = Function(fs, lambda x: x[0] + x[1] ** 2, name='f')
u = Function(fs, name='u')
u_solver = bvp(poisson, bcs)(u, f)
u_solver.solve()
plot_colormap(u)
(<Figure size 640x480 with 2 Axes>, <Axes: xlabel='$x$', ylabel='$y$'>)
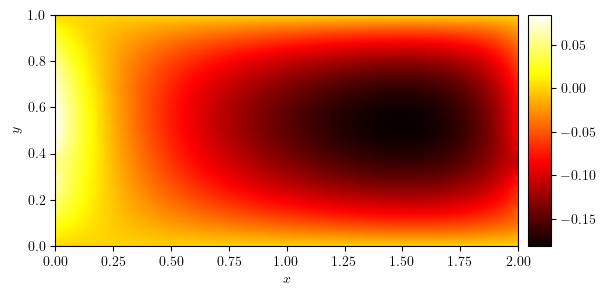
x_axis, ux, y_value = cross_section(u, 'y', 0.5)
plot_line((x_axis, ux), x_label='$x$', y_label=f'${u.name}(y={y_value:.2f})$')
(<Figure size 640x480 with 1 Axes>, <Axes: xlabel='$x$', ylabel='$u(y=0.50)$'>)
Cross-periodicity#
\[c(x,y=L_y)=c(x=0,y)\]
\[c(x=0,y)=c(x=L_x,y)=0\]
def upper_to_left(x: np.ndarray) -> np.ndarray:
return np.vstack((np.zeros_like(x[0]), (Ly / Lx) * x[0]))
f = Function(fs, lambda x: 1.0 + x[0] + x[1] ** 2, name='f')
bcs = BoundaryConditions(
('periodic', boundary['upper'], upper_to_left),
('dirichlet', boundary['right', 'lower'], 0.0),
)
u = Function(fs, name='u')
petsc = OptionsPETSc('preonly', 'lu', 1e-12, 1e-14)
u_solver = bvp(poisson, bcs, petsc)(u, f)
u_solver.solve()
plot_colormap(u, use_cache=False)
(<Figure size 640x480 with 2 Axes>, <Axes: xlabel='$x$', ylabel='$y$'>)
The Kernel crashed while executing code in the current cell or a previous cell.
Please review the code in the cell(s) to identify a possible cause of the failure.
Click <a href='https://aka.ms/vscodeJupyterKernelCrash'>here</a> for more info.
View Jupyter <a href='command:jupyter.viewOutput'>log</a> for further details.