Time-dependent boundary conditions#
-->See advection-diffusion-reaction demo for definition of initial boundary value problem.
import numpy as np
from lucifex.mesh import interval_mesh
from lucifex.fdm import (
BE, finite_difference_order,
FiniteDifference, FunctionSeries, ConstantSeries,
)
from lucifex.sim import run, Simulation
from lucifex.fem import Constant
from lucifex.solver import ibvp, evaluation, BoundaryConditions
from lucifex.viz import plot_line, create_animation, save_figure, display_animation
from lucifex.pde.diffusion import diffusion_reaction
def sine_wave(t, eps, omega):
return eps * np.sin(omega * float(t))
def create_simulation(
Lx: float,
Nx: int,
dt: float,
eps: float,
omega: float,
bc_type: str,
D_diff: FiniteDifference,
D_reac: FiniteDifference,
) -> Simulation:
order = finite_difference_order(D_diff.order, D_reac.order)
store = 1
mesh = interval_mesh(Lx, Nx)
t = ConstantSeries(mesh, name='t', ics=0.0)
dt = Constant(mesh, dt, name='dt')
uB = ConstantSeries(mesh, order=order, store=store, ics=sine_wave(0.0, eps, omega))
if bc_type == 'dirichlet':
uB.name = 'uD'
bcs = BoundaryConditions(
('dirichlet', lambda x: x[0], uB[1]),
('dirichlet', lambda x: x[0] - Lx, 0.0),
)
elif bc_type == 'neumann':
uB.name = 'uN'
bcs = BoundaryConditions(
('neumann', lambda x: x[0], -uB[1]),
('dirichlet', lambda x: x[0] - Lx, 0.0),
)
else:
raise ValueError
uB_solver = evaluation(
uB,
sine_wave,
future=True,
)(t[0] + dt, eps, omega)
u = FunctionSeries((mesh, 'P', 1), 'u', order, store, ics=0.0)
r = 1 - u
u_solver = ibvp(diffusion_reaction, bcs=bcs)(u, dt, 1, D_diff, r, D_reac)
return Simulation([uB_solver, u_solver], t, dt)
Time-dependent Dirichlet boundary condition#
\[\begin{split}
\mathbb{S}
\begin{cases}
\Omega = [0, L_x] \\
u_{\text{D}}(x=0, t)=\epsilon\sin(\omega t) \\
u_{\text{D}}(x=L_x)=0
\end{cases}
\end{split}\]
Lx = 1.0
Nx = 100
dt = 0.01
eps = 0.1
omega = 20
simulation = create_simulation(Lx, Nx, dt, eps, omega, 'dirichlet', BE, BE)
n_stop = 50
run(simulation, n_stop)
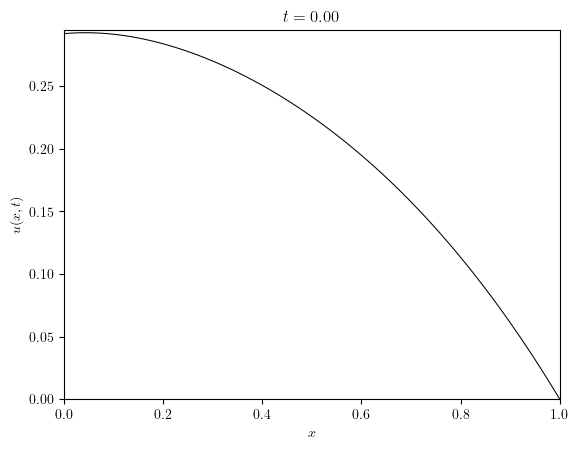
u = simulation['u']
u_min = np.min([np.min(dofs) for dofs in u.dofs_series])
u_max = np.max([np.max(dofs) for dofs in u.dofs_series])
title_series = [f'$t={t:.2f}$' for t in u.time_series]
anim = create_animation(
plot_line,
y_lims=(u_min, u_max),
x_label='$x$',
y_label='$u(x,t)$'
)(u.series, title=title_series)
anim_path = save_figure('dirichlet_u(t)', get_path=True)(anim)
display_animation(anim_path)
The Kernel crashed while executing code in the current cell or a previous cell.
Please review the code in the cell(s) to identify a possible cause of the failure.
Click <a href='https://aka.ms/vscodeJupyterKernelCrash'>here</a> for more info.
View Jupyter <a href='command:jupyter.viewOutput'>log</a> for further details.
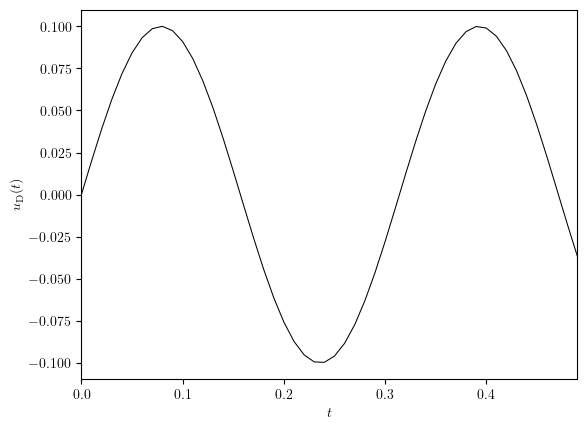
uD = simulation['uD']
fig, ax = plot_line((uD.time_series, uD.value_series), x_label='$t$', y_label='$u_{\mathrm{D}}(t)$')
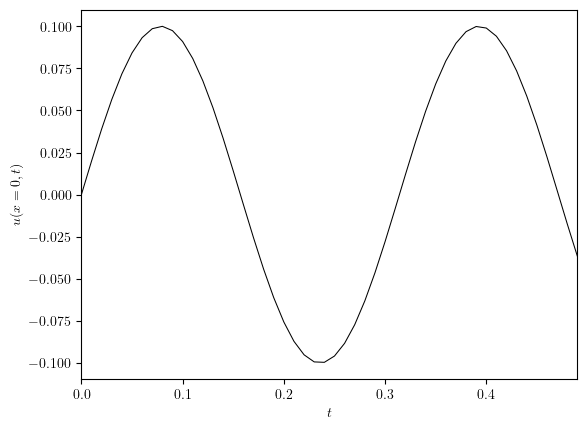
u = simulation['u']
uD_num = [dofs[0] for dofs in u.dofs_series]
fig, ax = plot_line((uD.time_series, uD_num), x_label='$t$', y_label='$u(x=0, t)$')
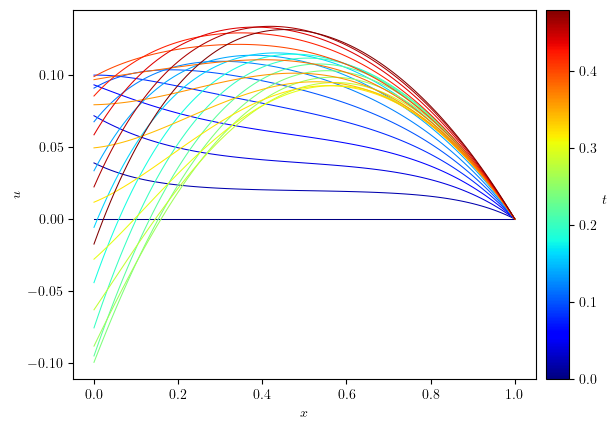
slc = slice(0, None, 2)
legend_labels=(min(u.time_series[slc]), max(u.time_series[slc]))
fig, ax = plot_line(u.series[slc], legend_labels, '$t$', cyc='jet', x_label='$x$', y_label='$u$')
save_figure('dirichlet_u(t)')
(<Figure size 640x480 with 2 Axes>, <Axes: xlabel='$x$', ylabel='$u$'>)
Time-dependent Neumann boundary condition#
\[\begin{split}
\mathbb{S}
\begin{cases}
\Omega = [0, L_x] \\
u_{\text{N}}(x=0, t)=\epsilon\sin(\omega t) \\
u_{\text{D}}(x=L_x)=0
\end{cases}
\end{split}\]
Lx = 1.0
Nx = 100
dt = 0.01
eps = 0.1
omega = 20
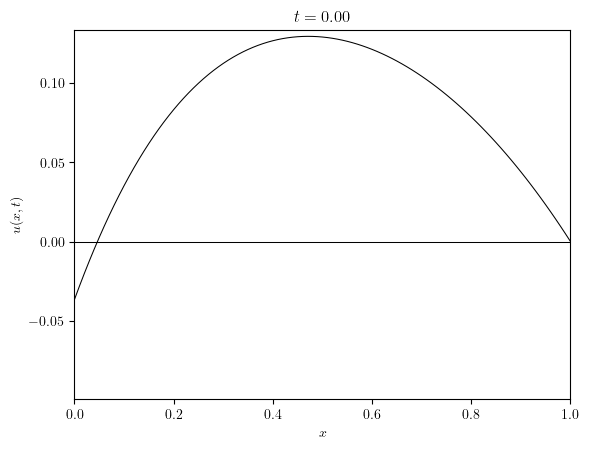
simulation = create_simulation(Lx, Nx, dt, eps, omega, 'neumann', BE, BE)
n_stop = 50
run(simulation, n_stop)
u = simulation['u']
u_min = np.min([np.min(dofs) for dofs in u.dofs_series])
u_max = np.max([np.max(dofs) for dofs in u.dofs_series])
title_series = [f'$t={t:.2f}$' for t in u.time_series]
anim = create_animation(
plot_line,
y_lims=(u_min, u_max),
x_label='$x$',
y_label='$u(x,t)$'
)(u.series, title=title_series)
anim_path = save_figure('neumann_u(t)', get_path=True)(anim)
display_animation(anim_path)